For clinicians
Dear Colleagues
Patients with intellectual disability, epilepsies and/or hyperphosphatasia (elevated alkaline phosphatase activity in the serum) and/or characteristic facial features a GPI-anchor deficiency should be suspected.
The diagnostic workflow for suspected GPIBDs contains
- analysis of exome or genome data (of parent index trio)
- prioritization of variant data based on facial image analysis by face2gene and phenotypic data (in HPO terms)
- functional characterization of pathogenic variants by flow cytometry on a freshly drawn blood sample in BCT
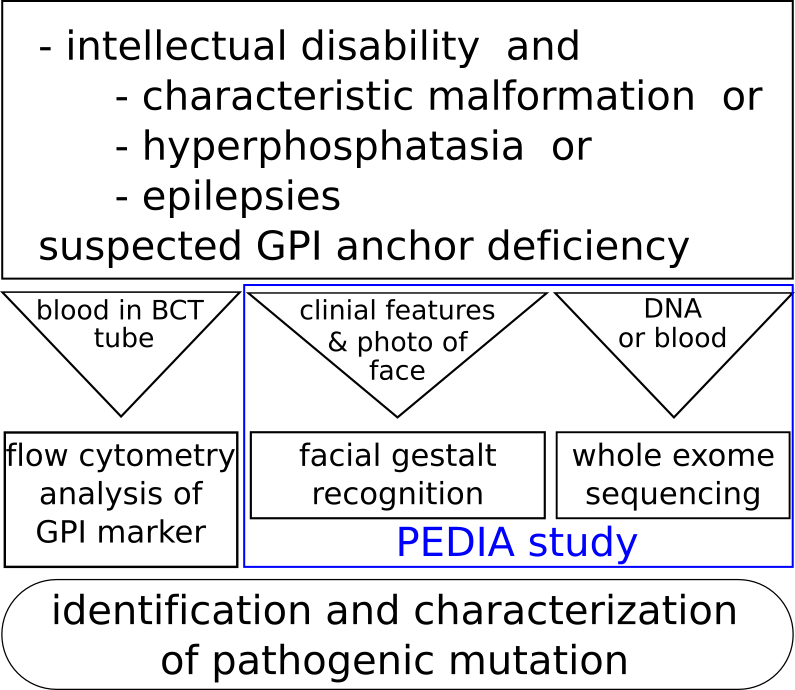
For such cases, we offer exome analysis including all known genes of the GPI-anchor synthesis on a research basis (GPI-gene panel diagnostics). For this analysis we need DNA (>3µg) or EDTA blood (>2ml).
Are you already familiar with Face2Gene CLINIC? We would highly appreciate if you submit your case via the LABS interface to UKBonn. This allows us to derive all the phenotypic data that we need for the bioinformatics prioritization automatically. Please contact us for more information.
If we cannot find any pathogenic mutations in the known genes we will analyse by flow cytometry whether the expression of GPI-anchored markers is reduced. For this analysis we need a fresh blood draw in a Cyto Chex tube. In any way you need to inform us before you send the sample as these samples have to be handled with care and the analysis has to be performed in a timely manner.
Please pay attention to the follow points when sending blood samples for flow cytometry analyses:
• Use the CytoChex BCT tubes for collection of fresh blood.
• Fill the tube with at least 2ml of fresh blood of the patient.
• Fill at least 2ml of fresh blood from an unrelated healthy individual for travel control.
• Make sure patient sample and travel control have an equal filling level in the tube.
• Use express delivery at 4°C to send the samples within 48 - 72 hours to:
Alexej Knaus
2G/603
Venusberg-Campus 1, House 11, 2nd floor
53127 Bonn
If you already have an NGS data set of an individual with a suspected GPI-anchor deficiency, e.g. a vcf file of an exome, you could also filter the data with the in silico gene panel at GeneTalk.
If we can confirm a GPI-anchor deficiency by flow cytometry we will add the individuals to a cohort with GPI-anchor deficiencies that do not have pathogenic mutations in the known GPI-anchor synthesis genes. We will sequence the exomes, genomes and transcriptomes of the individuals and their parents. We will analyze the sequence data of this cohort to identify new genes that are involved in the GPI-anchor synthesis and maturation.